Repository of scripts used by IRIDIA members
Max's scripts
When testing different algorithms on some combinatorial optimization problem, I usually organize the data on my machine according to a same schema. Basically I create the following structure of directories to store sources, problem instances, output data, executables and scripts.
+ main_project_folder | |-+ bin (contains the executables of the algorithms) | |-+ instances (contains the problem instances) | |-+ out (contains the outputs of the trial) | |-+ sh (contains the script to launch the experiments) | |-+ src (contains the sources of the algorithms)
Here I show you as an example what I did for the experiments on Parallel ACO for QAP:
mmanfrin@majorana:~/Parallel-QAP$ ls -al total 40 drwxr-xr-x 8 mmanfrin mmanfrin 4096 2006-07-06 15:11 . drwxr-xr-x 9 mmanfrin mmanfrin 4096 2006-08-14 12:29 .. drwxr-xr-x 2 mmanfrin mmanfrin 4096 2006-07-06 22:15 bin drwxr-xr-x 2 mmanfrin mmanfrin 4096 2006-08-09 17:02 instances drwxr-xr-x 16 mmanfrin mmanfrin 4096 2006-06-26 16:33 instances.all -rw-r--r-- 1 mmanfrin mmanfrin 40 2006-07-06 15:11 lamhost-group1.txt -rw-r--r-- 1 mmanfrin mmanfrin 40 2006-07-06 15:11 lamhost-group2.txt drwxr-xr-x 41 mmanfrin mmanfrin 4096 2006-07-07 10:55 out drwxr-xr-x 41 mmanfrin mmanfrin 4096 2006-08-09 17:00 sh drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-06-23 12:29 src
The bin directory contains links to the executables of the various algorithm:
mmanfrin@majorana:~/Parallel-QAP$ ls -al bin total 152 drwxr-xr-x 2 mmanfrin mmanfrin 4096 2006-07-06 22:15 . drwxr-xr-x 8 mmanfrin mmanfrin 4096 2006-07-06 15:11 .. lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:06 fc.1.x.10 -> ../src/MPI/sync/multicolony/fully-connected/FC.1.x.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:07 fc.2.6.10 -> ../src/MPI/sync/multicolony/fully-connected/FC.2.6.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:07 fc.2.7.10 -> ../src/MPI/sync/multicolony/fully-connected/FC.2.7.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:07 fc.2.8.10 -> ../src/MPI/sync/multicolony/fully-connected/FC.2.8.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:07 fc.2.9.10 -> ../src/MPI/sync/multicolony/fully-connected/FC.2.9.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:12 fc.3.6.10 -> ../src/MPI/sync/multicolony/fully-connected/FC.3.6.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:12 fc.3.7.10 -> ../src/MPI/sync/multicolony/fully-connected/FC.3.7.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:12 fc.3.8.10 -> ../src/MPI/sync/multicolony/fully-connected/FC.3.8.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:12 fc.3.9.10 -> ../src/MPI/sync/multicolony/fully-connected/FC.3.9.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:13 hc.1.x.10 -> ../src/MPI/sync/multicolony/hypercube/3D/HC.1.x.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:13 hc.2.6.10 -> ../src/MPI/sync/multicolony/hypercube/3D/HC.2.6.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:13 hc.2.7.10 -> ../src/MPI/sync/multicolony/hypercube/3D/HC.2.7.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:13 hc.2.8.10 -> ../src/MPI/sync/multicolony/hypercube/3D/HC.2.8.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:13 hc.2.9.10 -> ../src/MPI/sync/multicolony/hypercube/3D/HC.2.9.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:14 hc.3.6.10 -> ../src/MPI/sync/multicolony/hypercube/3D/HC.3.6.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:14 hc.3.7.10 -> ../src/MPI/sync/multicolony/hypercube/3D/HC.3.7.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:14 hc.3.8.10 -> ../src/MPI/sync/multicolony/hypercube/3D/HC.3.8.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:14 hc.3.9.10 -> ../src/MPI/sync/multicolony/hypercube/3D/HC.3.9.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:08 rw.1.x.10 -> ../src/MPI/sync/multicolony/fully-connected/RW.1.x.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:08 rw.2.6.10 -> ../src/MPI/sync/multicolony/fully-connected/RW.2.6.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:08 rw.2.7.10 -> ../src/MPI/sync/multicolony/fully-connected/RW.2.7.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:11 rw.2.8.10 -> ../src/MPI/sync/multicolony/fully-connected/RW.2.8.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:11 rw.2.9.10 -> ../src/MPI/sync/multicolony/fully-connected/RW.2.9.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:11 rw.3.6.10 -> ../src/MPI/sync/multicolony/fully-connected/RW.3.6.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:11 rw.3.7.10 -> ../src/MPI/sync/multicolony/fully-connected/RW.3.7.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:11 rw.3.8.10 -> ../src/MPI/sync/multicolony/fully-connected/RW.3.8.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 65 2006-07-06 22:12 rw.3.9.10 -> ../src/MPI/sync/multicolony/fully-connected/RW.3.9.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 37 2006-06-26 11:23 seq -> ../src/Sequential/MMASQAP/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:14 ur.1.x.10 -> ../src/MPI/sync/multicolony/linear-array/UR.1.x.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:14 ur.2.6.10 -> ../src/MPI/sync/multicolony/linear-array/UR.2.6.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:15 ur.2.7.10 -> ../src/MPI/sync/multicolony/linear-array/UR.2.7.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:15 ur.2.8.10 -> ../src/MPI/sync/multicolony/linear-array/UR.2.8.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:15 ur.2.9.10 -> ../src/MPI/sync/multicolony/linear-array/UR.2.9.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:15 ur.3.6.10 -> ../src/MPI/sync/multicolony/linear-array/UR.3.6.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:15 ur.3.7.10 -> ../src/MPI/sync/multicolony/linear-array/UR.3.7.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:15 ur.3.8.10 -> ../src/MPI/sync/multicolony/linear-array/UR.3.8.10/bin/mmasqap lrwxrwxrwx 1 mmanfrin mmanfrin 62 2006-07-06 22:15 ur.3.9.10 -> ../src/MPI/sync/multicolony/linear-array/UR.3.9.10/bin/mmasqap
The instances directory contains the problem instances to solve:
mmanfrin@majorana:~/Parallel-QAP$ ls -al instances total 588 drwxr-xr-x 2 mmanfrin mmanfrin 4096 2006-08-09 17:02 . drwxr-xr-x 8 mmanfrin mmanfrin 4096 2006-07-06 15:11 .. -rw-r--r-- 1 mmanfrin mmanfrin 33052 2006-06-23 12:28 lipa80a.dat -rw-r--r-- 1 mmanfrin mmanfrin 41900 2006-06-23 12:28 lipa90a.dat -rw-r--r-- 1 mmanfrin mmanfrin 61012 2006-06-23 12:28 sko100a.dat -rw-r--r-- 1 mmanfrin mmanfrin 61014 2006-06-23 12:28 sko100e.dat -rw-r--r-- 1 mmanfrin mmanfrin 40189 2006-06-23 12:28 sko81.dat -rw-r--r-- 1 mmanfrin mmanfrin 49514 2006-06-23 12:28 sko90.dat -rw-r--r-- 1 mmanfrin mmanfrin 60217 2006-06-23 12:28 tai100a.dat -rw-r--r-- 1 mmanfrin mmanfrin 120117 2006-06-23 12:28 tai100b.dat -rw-r--r-- 1 mmanfrin mmanfrin 38574 2006-06-23 12:28 tai80a.dat
The out directory contains a directory for each algorithm to be tested. Each directory contains a directory for each problem instance to be solved:
mmanfrin@majorana:~/Parallel-QAP$ ls -al out/ total 176 drwxr-xr-x 41 mmanfrin mmanfrin 4096 2006-07-07 10:55 . drwxr-xr-x 8 mmanfrin mmanfrin 4096 2006-07-06 15:11 .. -rwxr-xr-x 1 mmanfrin mmanfrin 11531 2006-07-07 10:55 create-dir.sh drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 fc.1.x.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 fc.2.6.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 fc.2.7.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 fc.2.8.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 fc.2.9.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 fc.3.6.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 fc.3.7.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 fc.3.8.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 fc.3.9.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 hc.1.x.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 hc.2.6.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 hc.2.7.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 hc.2.8.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 hc.2.9.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 hc.3.6.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 hc.3.7.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 hc.3.8.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 hc.3.9.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-04 17:28 pir drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 rw.1.x.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 rw.2.6.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 rw.2.7.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 rw.2.8.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 rw.2.9.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 rw.3.6.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 rw.3.7.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 rw.3.8.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 rw.3.9.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-04 17:31 seq drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-04 17:32 seq2 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 ur.1.x.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 ur.2.6.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 ur.2.7.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 ur.2.8.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 ur.2.9.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 ur.3.6.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 ur.3.7.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 ur.3.8.10 drwxr-xr-x 4 mmanfrin mmanfrin 4096 2006-07-07 10:55 ur.3.9.10
The bin directory contains links to the executables of the various algorithm:
The bin directory contains links to the executables of the various algorithm:
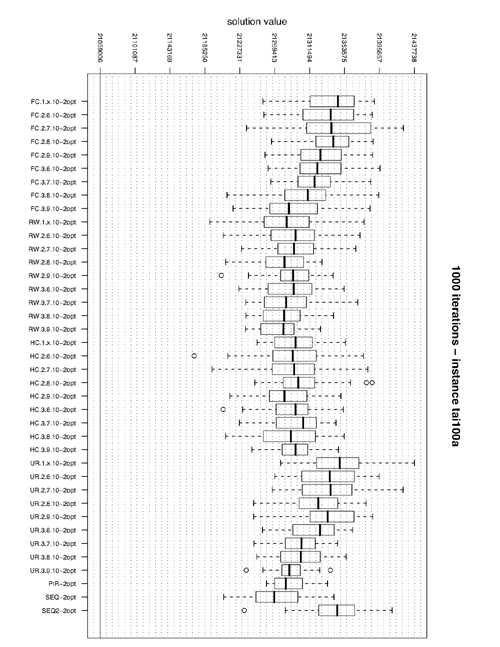
Boxplot of solution values by algorithm
I organize the data in the following way:
in the file optimal_values.txt I put the best-known solution values for the problem instances I'm testing
cat optimal_values.txt instance optimum sko100a 152002 sko100e 149150 tai100a 21059006 tai100b 1185996137
in the files algorithm_factors_instance-size_cut-time.txt I record the history of the search of the algo for the instance
head sequential_2-opt_100_8000.txt idalgo topo schema ls type cpu_id instance try best time iteration SEQ-2opt SEQ SEQ 2 Seq 0 sko100a 1 155468 0.31 1 SEQ-2opt SEQ SEQ 2 Seq 0 sko100a 1 155390 0.31 1 SEQ-2opt SEQ SEQ 2 Seq 0 sko100a 1 155000 0.31 1 SEQ-2opt SEQ SEQ 2 Seq 0 sko100a 1 154934 0.64 2 SEQ-2opt SEQ SEQ 2 Seq 0 sko100a 1 154608 0.97 3 SEQ-2opt SEQ SEQ 2 Seq 0 sko100a 1 153958 1.27 4 SEQ-2opt SEQ SEQ 2 Seq 0 sko100a 1 153750 1.93 6 SEQ-2opt SEQ SEQ 2 Seq 0 sko100a 1 153720 1.93 6 SEQ-2opt SEQ SEQ 2 Seq 0 sko100a 1 153634 2.57 8
In order to produce boxplots like the one above you can look at the R source used to produce it.
optimal_values<-read.table("optimal_values_100.txt",header=TRUE)
resPIR2OPT<-read.table("parallel_independent_2-opt_100_100.txt",header=TRUE)
resSEQ2OPT<-read.table("sequential_2-opt_100_800.txt",header=TRUE)
resSEQ22OPT<-read.table("sequential2_2-opt_100_100.txt",header=TRUE)
resFC1x102OPT<-read.table("fc.1.x.10_2-opt_100_100.txt",header=TRUE)
resFC26102OPT<-read.table("fc.2.6.10_2-opt_100_100.txt",header=TRUE)
resFC27102OPT<-read.table("fc.2.7.10_2-opt_100_100.txt",header=TRUE)
resFC28102OPT<-read.table("fc.2.8.10_2-opt_100_100.txt",header=TRUE)
resFC29102OPT<-read.table("fc.2.9.10_2-opt_100_100.txt",header=TRUE)
resFC36102OPT<-read.table("fc.3.6.10_2-opt_100_100.txt",header=TRUE)
resFC37102OPT<-read.table("fc.3.7.10_2-opt_100_100.txt",header=TRUE)
resFC38102OPT<-read.table("fc.3.8.10_2-opt_100_100.txt",header=TRUE)
resFC39102OPT<-read.table("fc.3.9.10_2-opt_100_100.txt",header=TRUE)
resHC1x102OPT<-read.table("hc.1.x.10_2-opt_100_100.txt",header=TRUE)
resHC26102OPT<-read.table("hc.2.6.10_2-opt_100_100.txt",header=TRUE)
resHC27102OPT<-read.table("hc.2.7.10_2-opt_100_100.txt",header=TRUE)
resHC28102OPT<-read.table("hc.2.8.10_2-opt_100_100.txt",header=TRUE)
resHC29102OPT<-read.table("hc.2.9.10_2-opt_100_100.txt",header=TRUE)
resHC36102OPT<-read.table("hc.3.6.10_2-opt_100_100.txt",header=TRUE)
resHC37102OPT<-read.table("hc.3.7.10_2-opt_100_100.txt",header=TRUE)
resHC38102OPT<-read.table("hc.3.8.10_2-opt_100_100.txt",header=TRUE)
resHC39102OPT<-read.table("hc.3.9.10_2-opt_100_100.txt",header=TRUE)
resRW1x102OPT<-read.table("rw.1.x.10_2-opt_100_100.txt",header=TRUE)
resRW26102OPT<-read.table("rw.2.6.10_2-opt_100_100.txt",header=TRUE)
resRW27102OPT<-read.table("rw.2.7.10_2-opt_100_100.txt",header=TRUE)
resRW28102OPT<-read.table("rw.2.8.10_2-opt_100_100.txt",header=TRUE)
resRW29102OPT<-read.table("rw.2.9.10_2-opt_100_100.txt",header=TRUE)
resRW36102OPT<-read.table("rw.3.6.10_2-opt_100_100.txt",header=TRUE)
resRW37102OPT<-read.table("rw.3.7.10_2-opt_100_100.txt",header=TRUE)
resRW38102OPT<-read.table("rw.3.8.10_2-opt_100_100.txt",header=TRUE)
resRW39102OPT<-read.table("rw.3.9.10_2-opt_100_100.txt",header=TRUE)
resUR1x102OPT<-read.table("ur.1.x.10_2-opt_100_100.txt",header=TRUE)
resUR26102OPT<-read.table("ur.2.6.10_2-opt_100_100.txt",header=TRUE)
resUR27102OPT<-read.table("ur.2.7.10_2-opt_100_100.txt",header=TRUE)
resUR28102OPT<-read.table("ur.2.8.10_2-opt_100_100.txt",header=TRUE)
resUR29102OPT<-read.table("ur.2.9.10_2-opt_100_100.txt",header=TRUE)
resUR36102OPT<-read.table("ur.3.6.10_2-opt_100_100.txt",header=TRUE)
resUR37102OPT<-read.table("ur.3.7.10_2-opt_100_100.txt",header=TRUE)
resUR38102OPT<-read.table("ur.3.8.10_2-opt_100_100.txt",header=TRUE)
resUR39102OPT<-read.table("ur.3.9.10_2-opt_100_100.txt",header=TRUE)
res<-rbind(resFC1x102OPT,resFC26102OPT,resFC27102OPT,resFC28102OPT,resFC29102OPT,resFC36102OPT,
resFC37102OPT,resFC38102OPT,resFC39102OPT,resRW1x102OPT,resRW26102OPT,resRW27102OPT,resRW28102OPT,
resRW29102OPT,resRW36102OPT,resRW37102OPT,resRW38102OPT,resRW39102OPT,resHC1x102OPT,resHC26102OPT,
resHC27102OPT,resHC28102OPT,resHC29102OPT,resHC36102OPT,resHC37102OPT,resHC38102OPT,resHC39102OPT,
resUR1x102OPT,resUR26102OPT,resUR27102OPT,resUR28102OPT,resUR29102OPT,resUR36102OPT,resUR37102OPT,
resUR38102OPT,resUR39102OPT,resPIR2OPT,resSEQ2OPT,resSEQ22OPT)
linstance<-levels(res$instance)
res.split<-split(1:nrow(res), list(res$instance, res$try, res$idalgo), drop=TRUE)
min.list <- lapply(res.split, function(x){
x[match(min(res$best[x]), res$best[x])]
})
min.vector <- unlist(min.list)
bestalgo<-res[min.vector,]
bestalgo.split <- split(1:nrow(bestalgo), bestalgo$instance, drop=TRUE)
for (i in (1:length(bestalgo.split)))
{
bestalgo.vector <- unlist(bestalgo.split[i])
bestalgo.temp <- bestalgo[bestalgo.vector,]
l<-split(bestalgo.temp$best,bestalgo.temp$idalgo)
epsfile=paste(linstance[i],"_100_nolim.eps",sep="")
postscript(file=epsfile,onefile=TRUE,horizontal=TRUE)
par(mar=c(5,5,5,3),cex.axis=0.7,las=2,mgp=c(4, 1, 0))
title_plot=paste("100 iterations - instance ",linstance[i],sep="")
boxplot(l,xlab="",ylab="solution value",names=c(levels(bestalgo$idalgo)),main=title_plot,
yaxt="n",ylim=c(optimal_values[optimal_values$instance==linstance[i],]$optimum,max(bestalgo.temp$best)))
axis(2, seq(from=optimal_values[optimal_values$instance==linstance[i],]$optimum,to=max(bestalgo.temp$best),length.out=10))
abline(h=optimal_values[optimal_values$instance==linstance[i],]$optimum)
# draw an orizontal line at the y-level of the best-know solution value
grid(nx=0, ny=55,col="gray5")
dev.off()
}
Things to improve
Use a separate text file that contains all the id data on an algorithm (idalgo, topo, schema, ls, type) to reduce the size of the results text files (for the QAP experiments I obtained something like 500MB of text results files, half of which are redundant data)
result.txt should look like: idalgo cpu_id instance try best time iteration ^^^^^^ ^^^^^^^^ algorithms.txt should look like: idalgo topo schema ls type number_of_cpus ^^^^^ instances.txt should look like: instance best-known-solution-value ^^^^^^^^